GUIDE
Basic function
Common in PC / Smartphone
User's Manual
 DOWNLOAD
DOWNLOAD
-
Member registrationPlease sign up as a member on the new account registration screen to use the app.*The password is very important information for you.
Please set a password that is difficult to guess by other people. -
LoginYou need to log in to the app to use.
enter your registered e-mail address and password. -
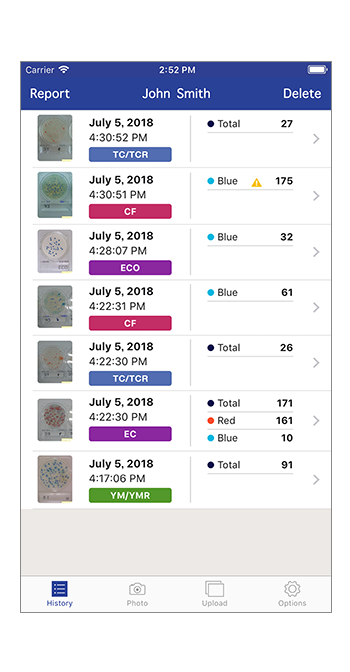
History of countYou can see the history of colony counts.*Any incorrect analysis triggers an error message.
Please see the list of reasons for errors. -
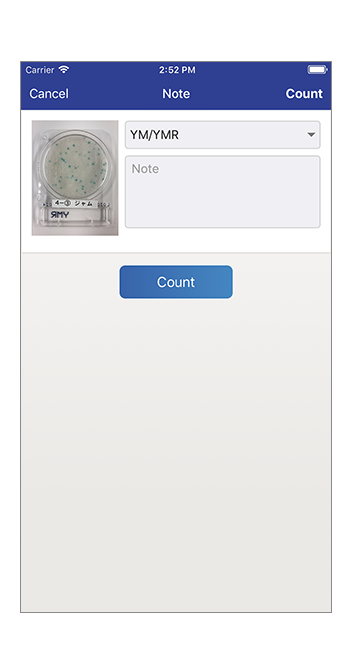
Photograph (for smartphone only)A camera function to take a picture of colonies on CompactDry™.
-
LibraryA function to count the number of colonies in saved images.*You can select a maximum of 10 images at once.
-
ReportAbility to report colony-counted images and results to specified email address in jpeg file and csv file.
-
DeleteFunction to delete saved colony-counted images data from cloud storage (aws).
STEP.1
Take a picture
Smartphone/digital camera
※The file format is jpeg or png only.

STEP.2
Upload the image
Smartphone/PC
※Simultaneous transmission number of Max is 10 files at a time.

STEP.3
Count the number of colonies
Automatic calculation of bacterial count.

Restrictions for input images in Colony Counter
No warranty of accuracy is given for the following images
-
1Image resolution of less than 800 pix x 1200 pix.
-
2The background color is not white.

-
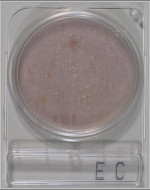
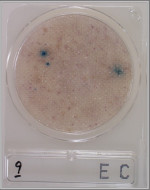
3Colored medium.

-
4A viscous sample is added.

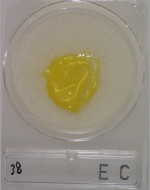

-
5Food residues are contained.

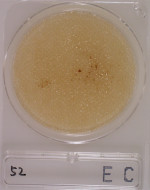

-
6Highly concentrated colonies.

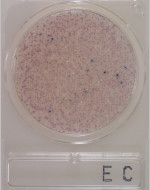

-
Other
- * The count result of "0" does not mean a negative result.
- * It may cause a count error of approximately 3% in some images.
- * Shimadzu Diagnostics takes no responsibility for the analysis results provided by this service.
- * Detection range is between 1–250 cfu/plate.
- * There are two types of misreads that can occur when using the @BactLAB™.
- @BactLAB™ Application did not detect all of the colonies specific to the CompactDry™ plate.
- The counter detected all the colonies but it categorized them incorrectly.